User Manual
Main Interface of MetaPro
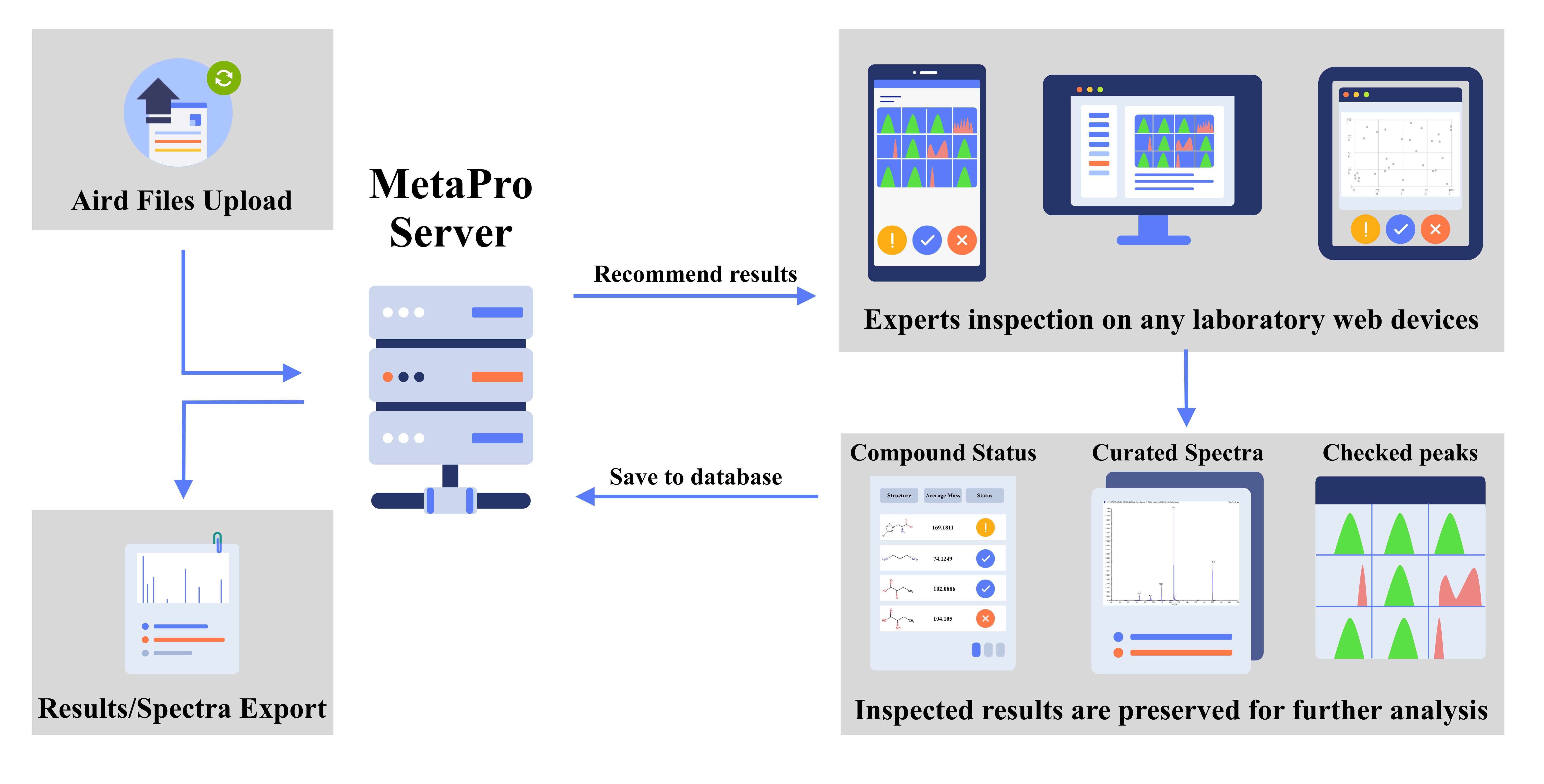
Framework of MetaPro
Demo website
1 General Information
1.1 Scope of MetaPro
Currently mass spectrometry instruments are widely used in metabolomics studies, but software in this filed usually have a lack of easily executed batch inspection and spectra curation ability. Besides, collaboration inconvenience on data processing also restrict analysis on efficiency.
MetaPro is a web-based stand-alone application for metabolomics MS data analysis, characterized by batch manual inspection and spectral library curation. MetaPro can perform accurate targeted compound detection on the input data and assure the result quality by batch manual inspection. Then, MetaPro can perform spectral library curation on the inspected results, which can improve the accuracy of further targeted analysis and promote the efficiency of manual inspection. The curated libraries and quantification results can be exported for further customized analysis.
1.2 System conception description
In MetaPro, we provide these main base compositon moduels:
Batch, Compound, Experiment, Library, Method, Overview, Project, Spectrum, Task.
The explanations for these base items are described below.
| Item | Explanation |
|---|---|
| Batch | A batch is composed of some experimental files. A project can have a few batches. |
| Compound | A compound refers to a specific metabolite whose name, ID, m/z, RT and other information are stored in the database. Selected compounds could be used for conducting targeted analysis. |
| Experiment | An experiment is a basic analytical unit in the MetaPro. It represents the mass spectrometry data obtained from one sample. The actual storage mode of an experiment is an Aird format file under the user-defined document folder. A few experiments can be divided into one batch. |
| Library | A library contains a few compounds. |
| Method | All parameters used in the analysis steps are extracted into a method. Different parameters composition could be saved into different methods for convenient query. |
| Overview | An overview is the storage of the analyzed results. By checking a overview, multiple quality control operations can be achieved. |
| Project | A project can have a few batches. Each batch has a few experiments. Project is the largest unit where experiments could be organized. |
| Spectrum | A spectrum is correlated to a specific compound. Instrument type, collision energy, ionization mode and other description information of the mass spectrometry characteristics are stored in a spectrum. |
| Task | Task refers to a metabolomics analysis process needed to be done or undertaken. It’s timely status and records could be seen in the task page. |
1.3 Data restrictions
MetaPro currently only supports DDA mode data. The input for MetaPro should be converted vendor files (Aird files). The compound library should be given to the application in a specific format.
1.4 Data safety
MetaPro is a stand-alone application that can be deployed on your personal computer or high-performance server. All data will be saved on your local devices and will not be uploaded.
1.5 Contact us
If you have any questions, you can or contact us through Github or csi@csibio.net.
2 Installation
2.1 System and hardware requirements
MetaPro is available for Windows 7 (or above), Linux and MacOS. The minimum and recommended system specifications are described in the following table.
| Specifications | Minimum | Recommended |
|---|---|---|
| CPU | Intel Core i5 10-gen or AMD R5 | Intel Core i9 12-gen or AMD R9 5900 |
| Hard drive | 50GB free space | 2x data set size |
| RAM | 16GB | 64GB(Core i9-10gen), 128GB(Core i9-12gen) |
2.2 Download
MetaPro
Installation packages for all the supported systems can be downloaded at GitHub.
AirdPro
MetaPro uses AirdPro for Vendor MS file conversion.
AirdPro (version 1.1.0.0) can be freely downloaded at GitHub.
Tutorial data
The tutorial data is a public metabolomics software benchmark dataset, published in Li, Zhucui, et al.
You can download the tutorial data at GoogleDrive and OneDrive.
The data contains 2 datasets:
- TripleTOF dataset
- 8 converted instrument data (16 files)
- 2 library (internal standard library, analyte library)
- QE HF dataset
- 10 converted instrument data (20 files)
- 2 library (internal standard library, analyte library)
2.3 One-click installation
Extract MetaPro zip file to any directory you want.
Here we use INSTALLATION_PATH to represent your installation path, for example C:\
Windows
Open INSTALLATION_PATH folder
start: Right click
MetaproStart.bat, run as administrator.Successful operation: The following operations appear in the terminal to indicate that the operation is successful:
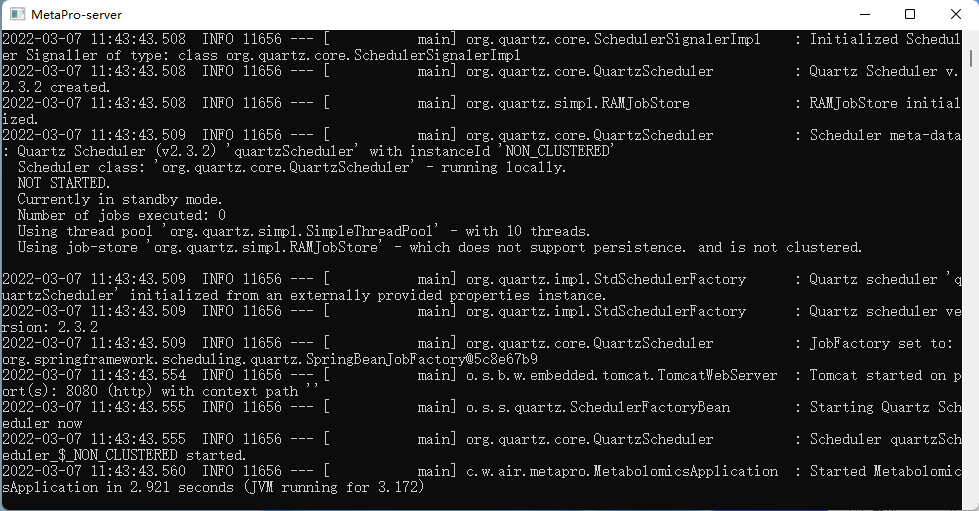 After successful startup, open a browser and connect to
After successful startup, open a browser and connect to localhost:8080stop: Right click
MetaproStop.bat, run as administratoruninstall: Stop MetaPro and delete the installation folder.
MacOS
- Open a terminal
cd INSTALLATION_PATH
cd MetaproForMac
- start:
bash MetaproStart.sh
Successful operation:
The following operations appear in the terminal to indicate that the operation is successful:
$ bash ./MetaproStart.sh
/Users/commands/MetaproForMac
********* Start Redis *********
/Users/commands/MetaproForMac/redis
********* Start MongoDB *********
/Users/commands/MetaproForMac/mongodb
********* Start Metapro *********
about to fork child process, waiting until server is ready for connections.
forked process: 94332
$ child process started successfully, parent exiting
After successful startup, open a browser and connect to localhost:8080
- stop:
bash MetaproStop.sh
- uninstall: Stop MetaPro and delete the installation folder.
Linux
- Open a terminal
cd INSTALLATION_PATH
cd MetaproForUbuntu
- start:
bash MetaproStart.sh
Successful operation:
The following operations appear in the terminal to indicate that the operation is successful:
$ bash ./MetaproStart.sh
/home/commands/MetaproForMac
********* Start Redis *********
/home/commands/MetaproForMac/redis
********* Start MongoDB *********
/home/commands/MetaproForMac/mongodb
********* Start Metapro *********
about to fork child process, waiting until server is ready for connections.
forked process: 94332
$ child process started successfully, parent exiting
After successful startup, open a browser and connect tolocalhost:8080
- stop:
bash MetaproStop.sh
- uninstall: Stop MetaPro and delete the installation folder.
After successful startup, open the browser to enter: localhost:8080, enter MetaPro interface.
Browsers support: Modern browsers and IE11: IE11, Edge, Firefox, Chrome, Safari, Opera
2.4 Configuration
There are two important parameters in the configuration of MetaPro. The analysis data directory and the result export directory.
Analysis data directory
All files that need to be analyzed by MetaPro need to be placed under the analysis data directory according to the rules in section 4.1.1. MetaPro can quickly import multiple batches of experiments by monitoring the converted files in the analysis data directory. All results that need to be exported by MetaPro will be exported to the result export directory.
Both directories are automatically set during the installation process. Users can modify the path according to their requirements.
Default directories are summarized as follows:
[Windows]
# default analysis data directory
C://metapro//data
# default result export directory
C://metapro//export
[MacOS]
# default analysis data directory
/Users/$USER/metapro/data
# default result export directory
/Users/$USER/metapro/export
[Ubuntu]
# default analysis data directory
/home/$USER/metapro/data
# default result export directory
/home/$USER/metapro/export
The way of changing default parameters is as follows:
- Open the
config.propertiesfile in the installation directory of MetaPro. - Change the value of
directoryandexport.directoryis the analysis data directory andexportis the result export directory. - Restart MetaPro.
2.5 Associated software
AirdPro
AirdPro (version 1.1.0.0) can be freely downloaded at GitHub.
MetaPro uses AirdPro (version 1.1.0.0) for instrument vendor file conversion. Instrument data should be converted to Aird format (.aird, .json) before being loaded into MetaPro. Aird is a computational-oriented data format for rapid access of MS data. By rearranging, compressing, and indexing spectra data, Aird can significantly speed up data reading, and thus improve the efficiency of data analysis and inspection.
AirdPro supports the vendor files including .raw, .wiff and .d formats.
AirdPro runs on Windows system, limited by the system requirement of vendor APIs.
3 Page Introduction
3.1 Project management page
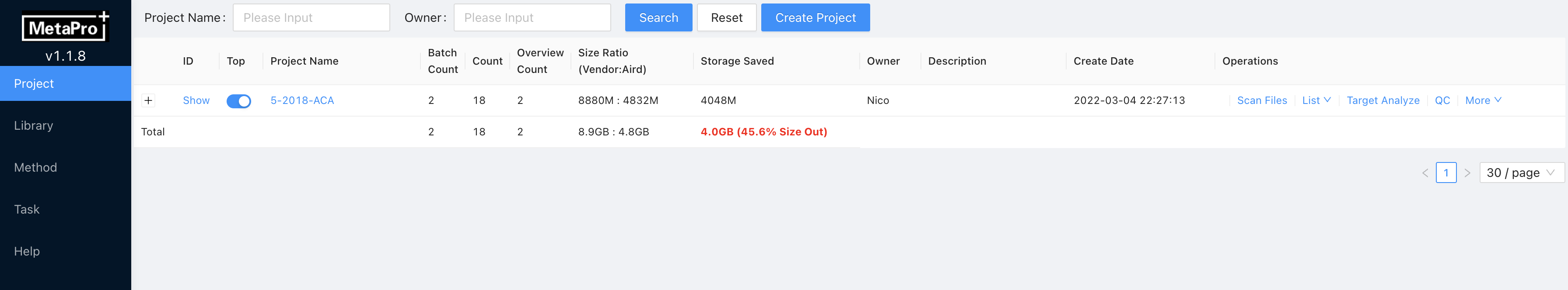
This is the first page that users will see when they start the system. Users can create new projects by clicking on the Create Project button in the top column. Then, input the name, owner, and description information of the new project on this page, where delete, update and search options are also available. The project management page shows all the users’ projects and their additional information, including batch numbers, experiment numbers, overview numbers, owner and create date. Storage space is shown with saved space because the use of Aird data format. To add experimental files into a project, users need to categorize all the Aird files into a folder having the same name with the created project. The folder should be put under the repository of MetaPro. This folder must contain several sub-folders, which are considered to be batches under this project. Aird files must be put under the sub-folders rather than the folder having the same name with the created project. Subsequently, these Aird files and batches information can be read into the system through clicking on the "scan file" button. During this process, their brief information will be stored. Meanwhile, batch number and experiment number are automatically modified according to the actual file arrangement under the repository. Also, Users can check extracted ion chromatogram (EIC) in each experiment after the index is created in the system. Clicking on the selected project will get access to its detailed information page, where its experiments information are fully displayed. Here, the experiment information is able to be updated, deleted, and modified.
3.2 Library management page
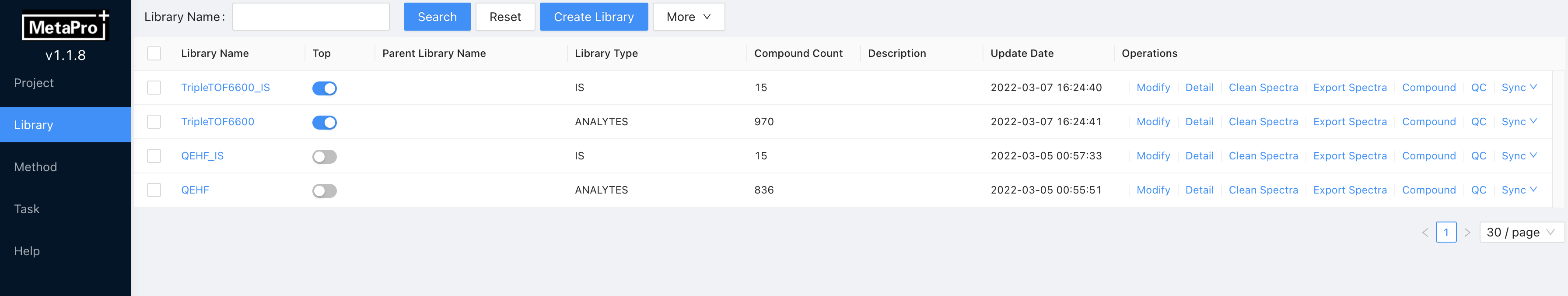
This page has the similar visualization hierarchical to the project management page. Creating, updating, modifying, deleting options toward compound libraries are available here. When creating a new library, MetaPro allows users to upload a formatted CSV file to fill the compounds information in this library. Clicking a library can check all the compounds that are included in this library.
3.3 Method management page
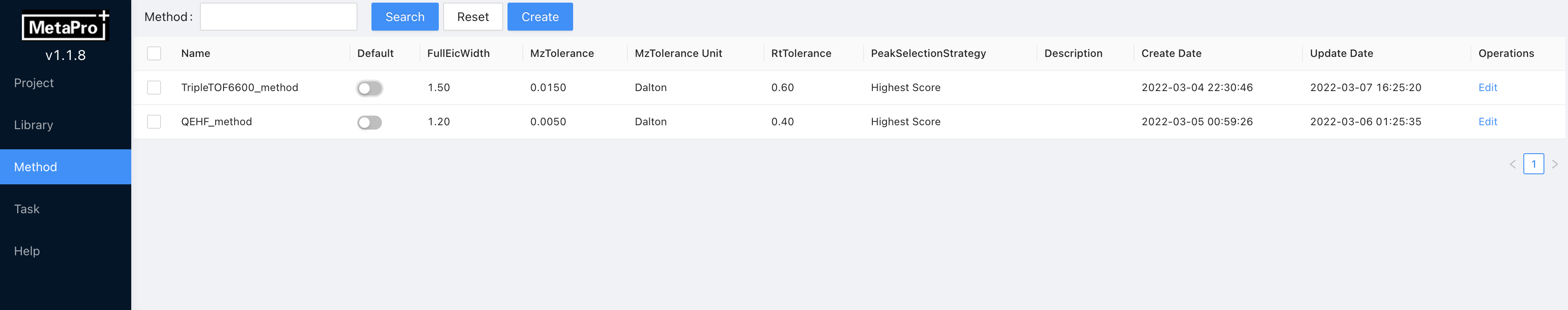
The contents shown here are the clustered parameters, which we call methods. When creating a new method, recommended parameters are preset in the method template. Noteworthy, created methods can be used to various projects, batches and experimental files. This design makes it easy to check parameter settings among different projects. Creating, viewing, deleting and modifying methods are operated on this page.
3.4 Task management page
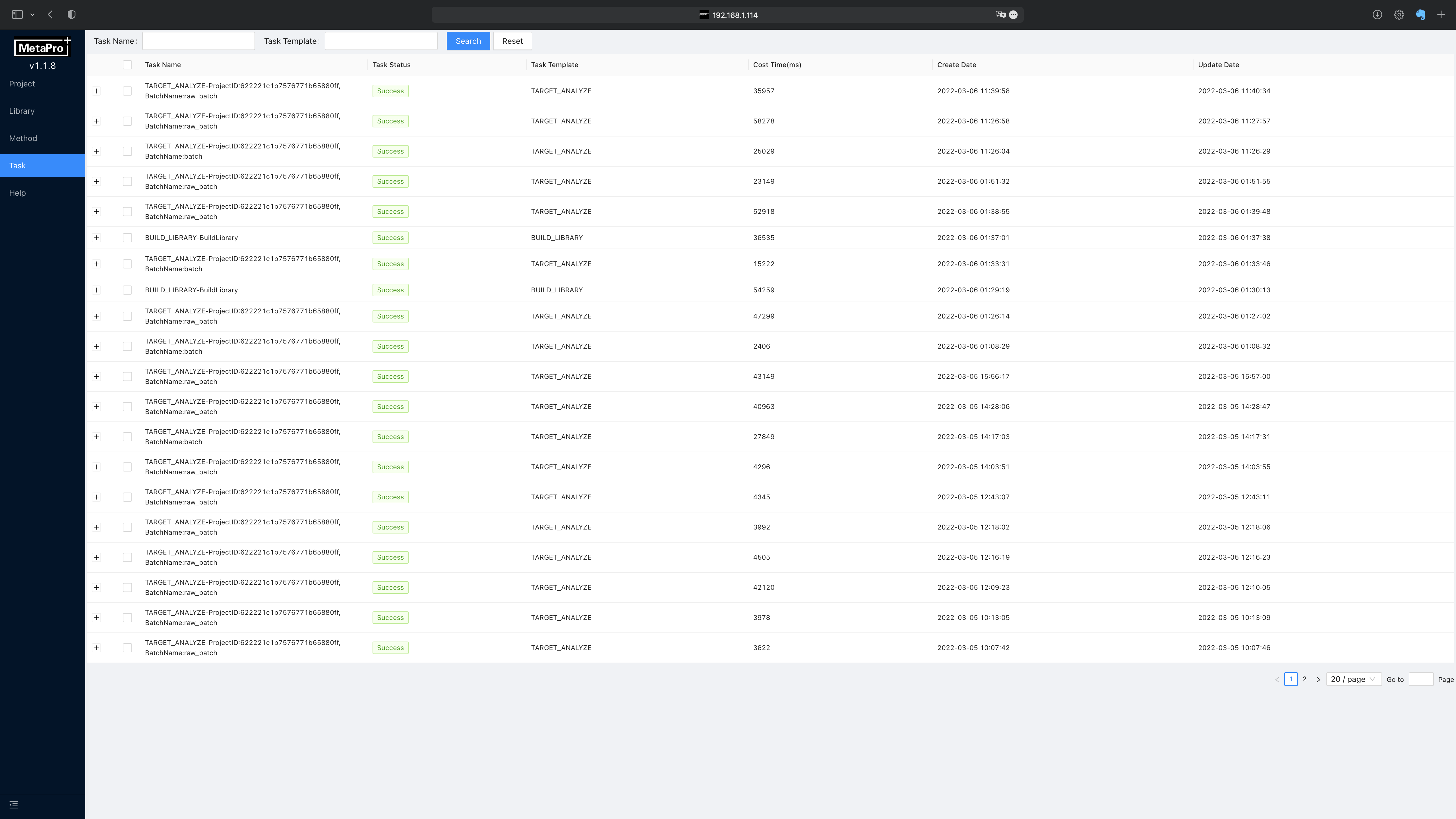
Task page presents the progress of BUILD_LIBRARY SCAN_AND_UPDATE_EXPERIMENTS. Users can check their execution status on this page. The execution log of each task can be viewed by clicking on the + button on each row. Time cost, created time, and updated dates are displayed for each task.
4 Step by step instructions
4.1 Data preparation
4.1.1 MS file preparation
Step 1: File Conversion
MetaPro supports MS data acquired with the data-dependent acquisition (DDA) method. Vendor MS data should be converted to Aird format (.aird & .json) before being loaded into MetaPro.
Introduction and download link of the file conversion software AirdPro is illustrated above in section 2.5.
Open AirdPro file converter Open AirdPro by running AirdPro.exe in the installation directory. Users can create a desktop shortcut for easy access.
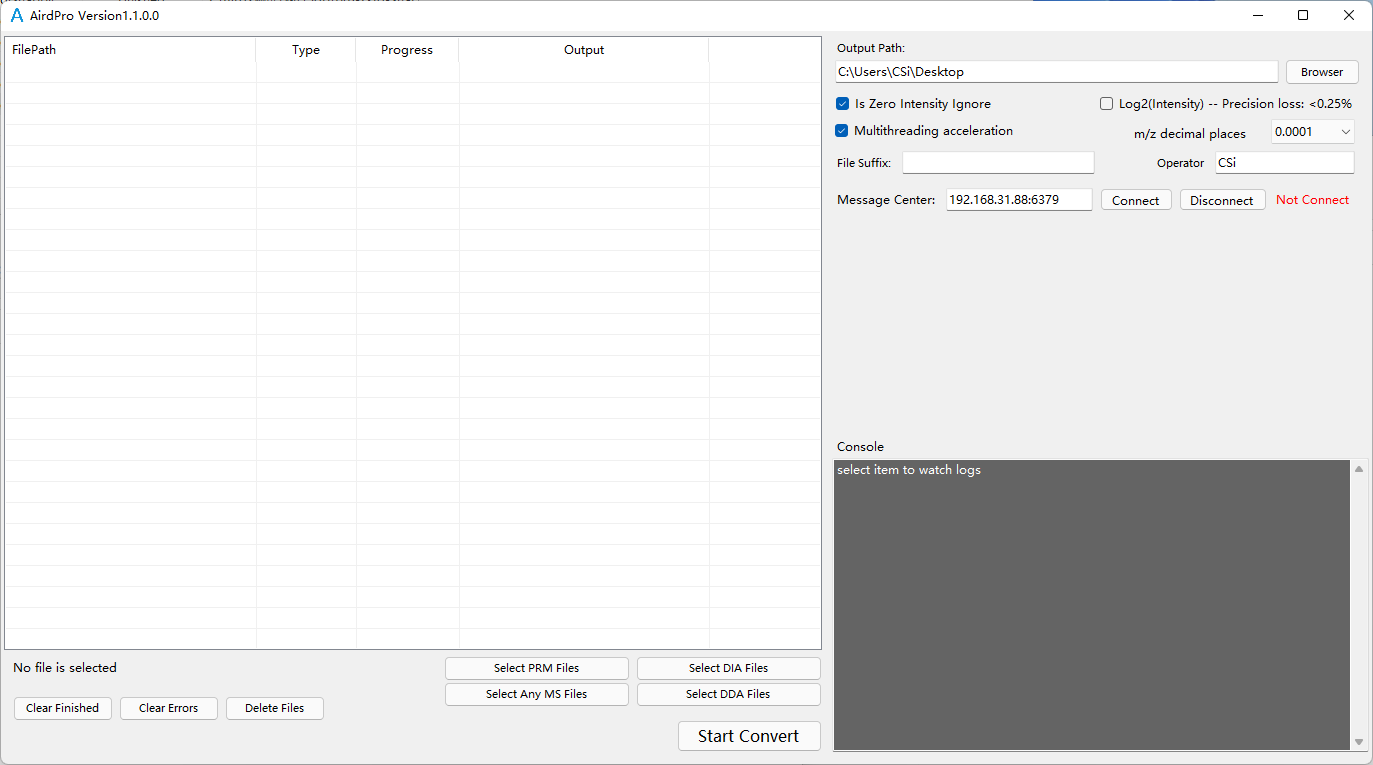
Select files for conversion in DDA mode
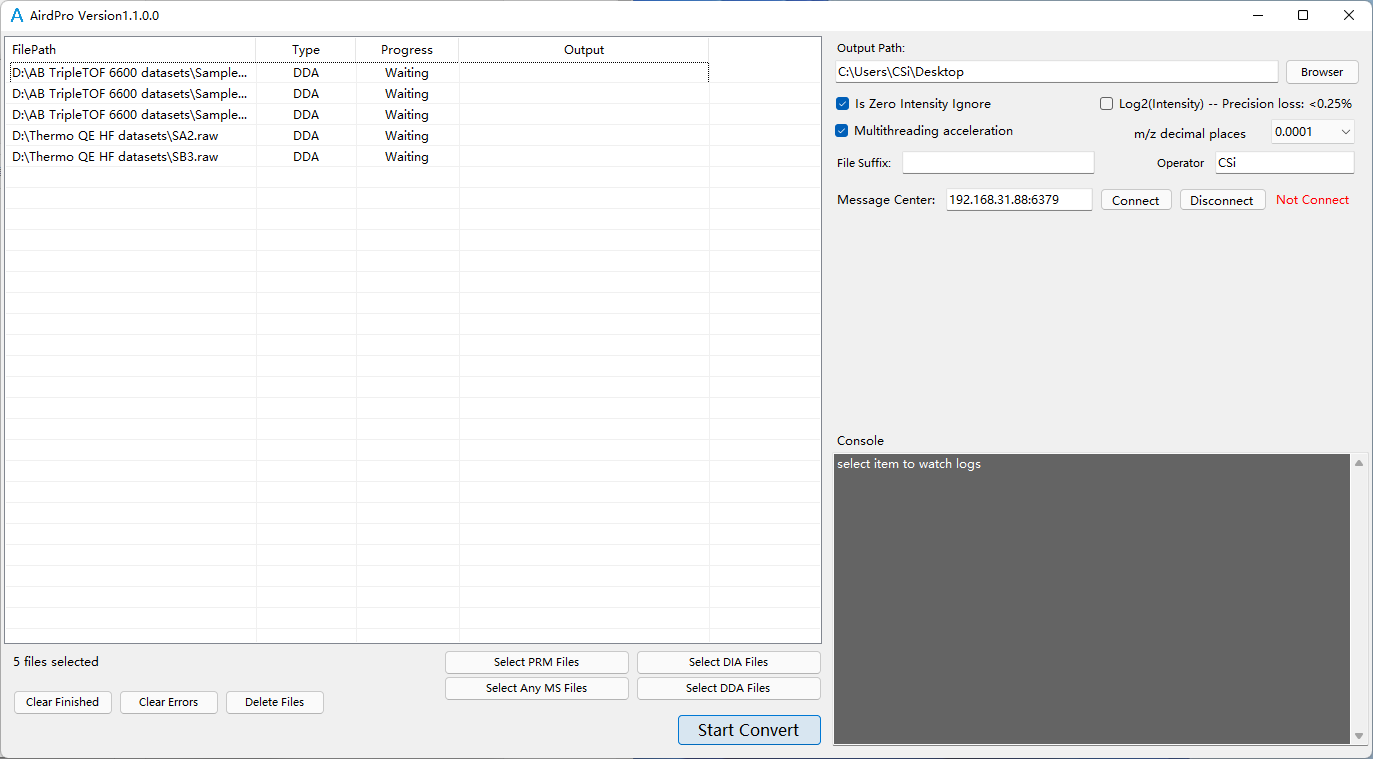
Select output folder
Start Convert After file conversion, each MS file is converted to two converted files, end with .aird and .json.
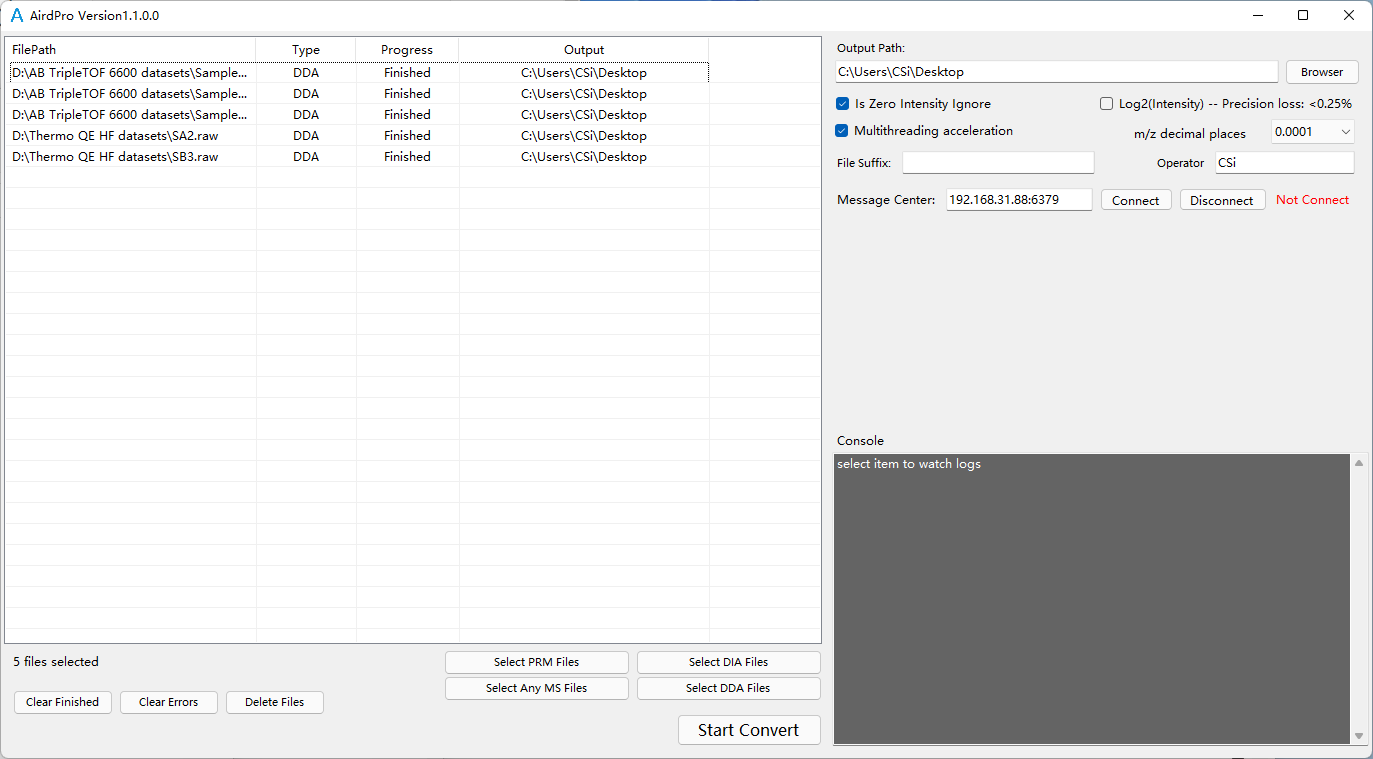 For example:
For example:# Thermo file Original: file_name.raw Converted: file_name.aird, file_name.json # SCIEX file Original: file_name.wiff, file_name.wiff.scan Converted: file_name.aird, file_name.jsonStep 2: File directory arrangement
MetaPro enables efficient multi-batch files import by scanning folder structure under the previously set data repository (section 2.4).
For example, if we set the data directory as
repository=/usr/local/dataThe folder structure of different projects needs to be arranged as follows:
/usr/local/data ├── project-1 ├── batch-1 ├── converted_file-1.aird ├── converted_file-1.json ├── converted_file-2.aird ├── converted_file-2.json ├── batch-2 ├── converted_file-3.aird ├── converted_file-3.json ├── converted_file-4.aird ├── converted_file-4.json ├── project-2 ├── ... ...The first-level directory under the scan directory is the project name. The second-level directory is the batch name. Converted aird files should be placed into batch folders.
- Note that: If your project has only ONE BATCH, you also need to create a batch folder in the project directory and put the converted files in the batch folder.
4.1.2 Library preparation
The Library should be a .txt/.tsv format table formatted in the following template. The first line at least needs to contain the mz and rt items. Other items are optional. Example compound library Analyte Library
Analyte Library
The analyte library is a list of compounds that you want to identify and quantify from the mass spectrometry data. MetaPro performs targeted data analysis based on this library.
If you do not have an analyte library, you can make your own analyte library based on the theoretical M/z and elution time of the compounds, download an open-source library, or modify analysis results of other software into an analyte library.
Internal Standard Library (Optional)
If you want to change the gradient of your experiment, for example, from 20min to 40min, the elution time of analytes would change and not elute at the RT value in the analyte library. In this circumstance, you can import a new analyte library with modified RT, or use an internal standard library.
- The internal standard library needs to be paired with the analyte library to correct the retention time of the analytes.
- The RT value in the internal standard library needs to be from the same gradient as the RT in the analyte library
- Compounds in internal standard library should have narrow and high-intensity chromatogram peaks.
- The compound number in the internal standard library should be about 10.
4.2 Start a targeted analysis task
4.2.1 Load file
Enter Project page, click
Create Projectat the top.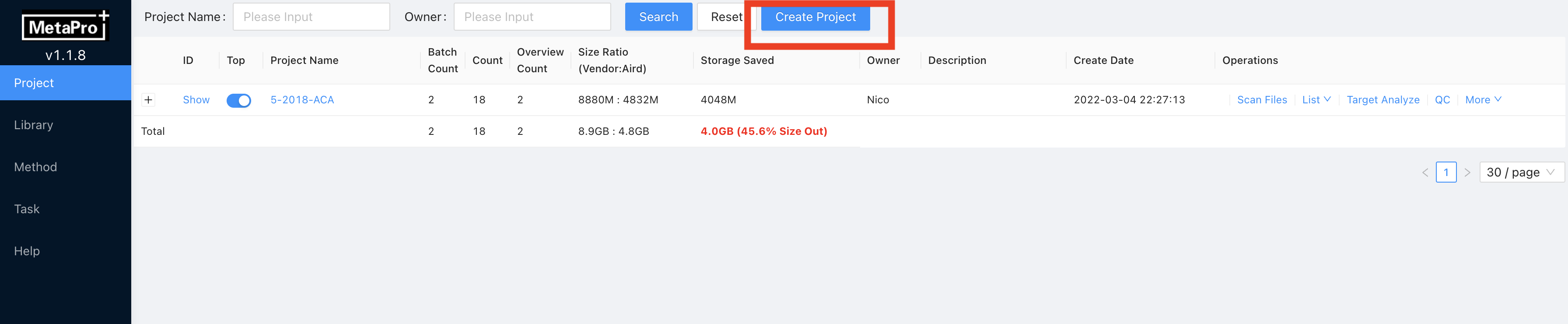 Then enter the
Then enter the Project Name. The project name should be the project name set in the File directory arrangement (section 4.1.1 Step 2). For example, if you set your project folder name as project-1, you need to name theProject Nameas project-1.OwnerandDescriptionare optional, fill in according to the demand.Click
Scan Filesto scan all converted Aird format files under the project file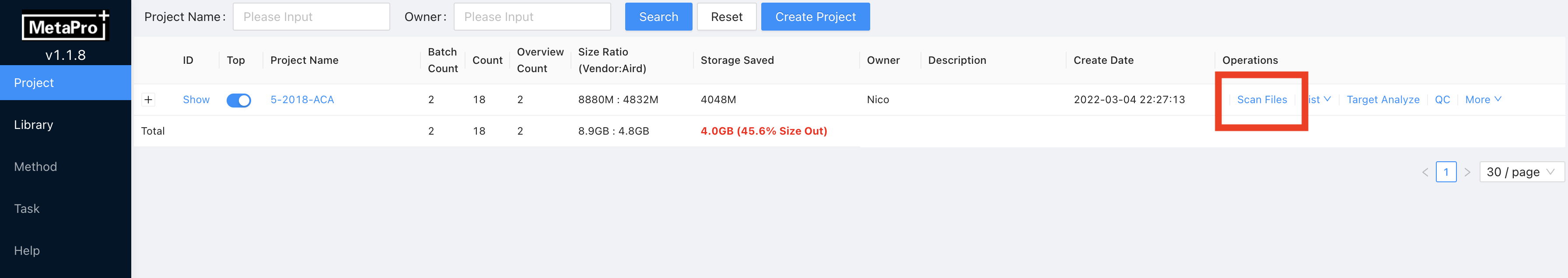 If MetaPro finds new files in the project folder, MetaPro will start to load the files and pop a message as follows:
If MetaPro finds new files in the project folder, MetaPro will start to load the files and pop a message as follows: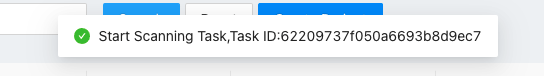
If find no new file or the folder is empty, MetaPro will pop up the following prompt box:
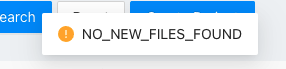
The file scanning progress can be find at the Task page. You can view the loaded files by
List → Samplesor single click on the project name. Sample list is presented as follows: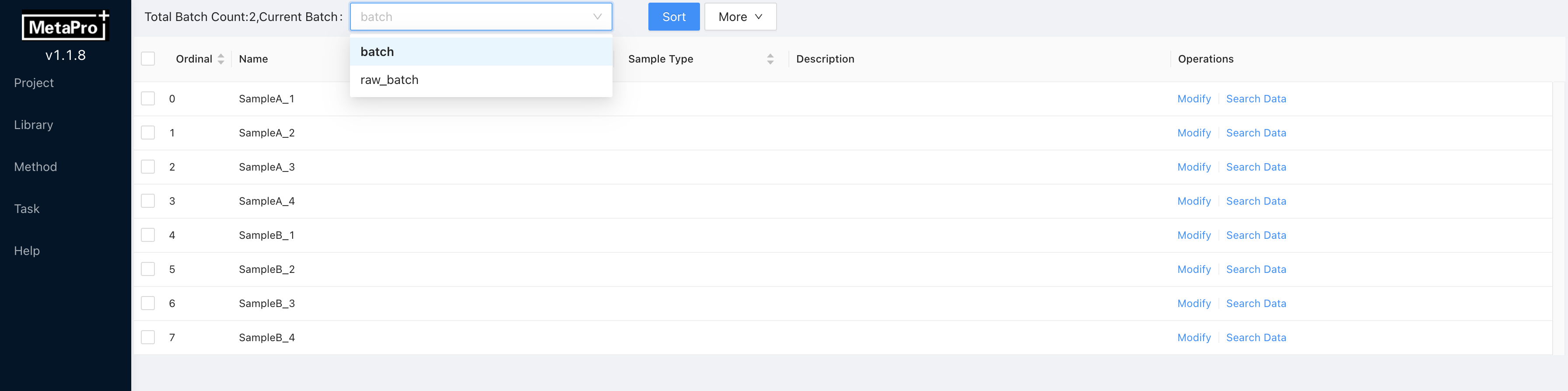
You can view the files in different batches by selecting Batch name at the top of this page.
4.2.2 Load library
Enter Library page, click Create Library at the top.
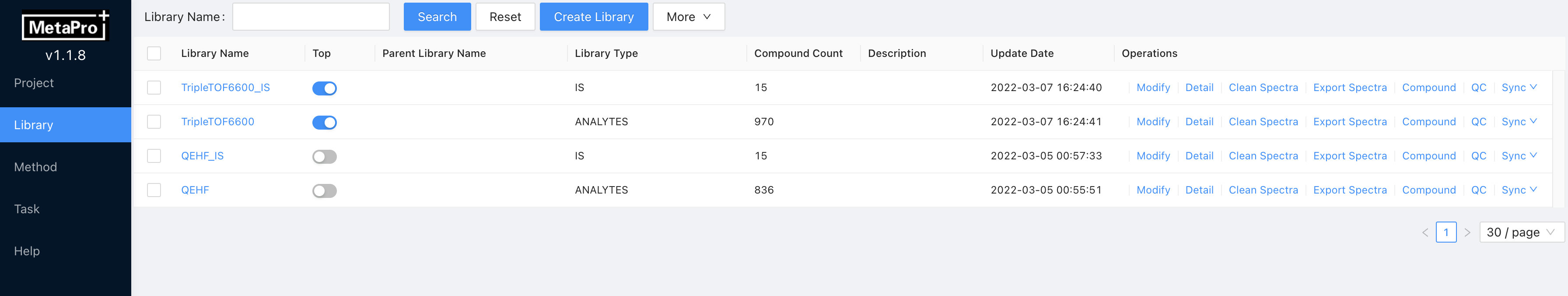
Input the Library Name, select the Library Type, and upload your library file at Upload File, click OK.
You must select a Library Type for this library. Select ANALYTES for the analyte library and IS for the internal standard library. If you forget to select library type, click Modify to set the library type before analyzing.
4.2.3 Set Method parameters
Enter Method page, click Create at the top.
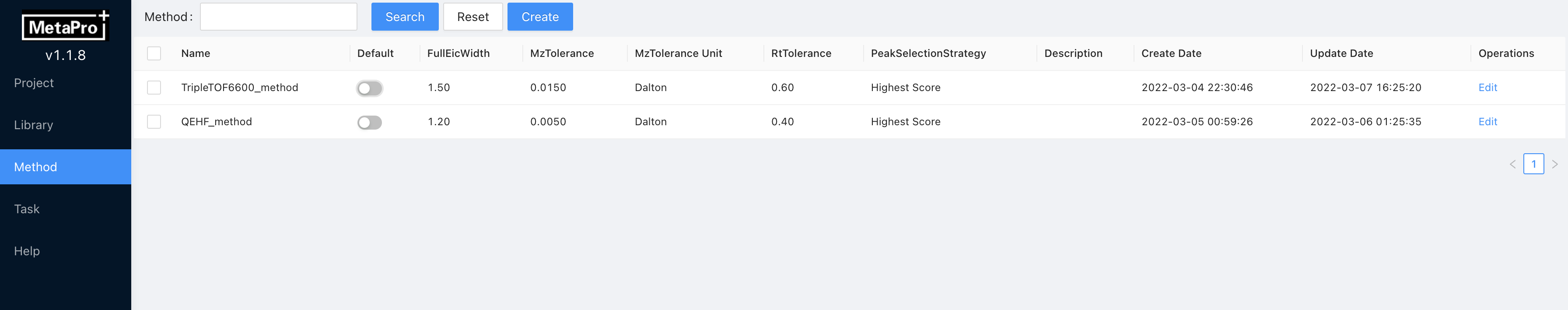
Input the Method Name, and set the method parameters.
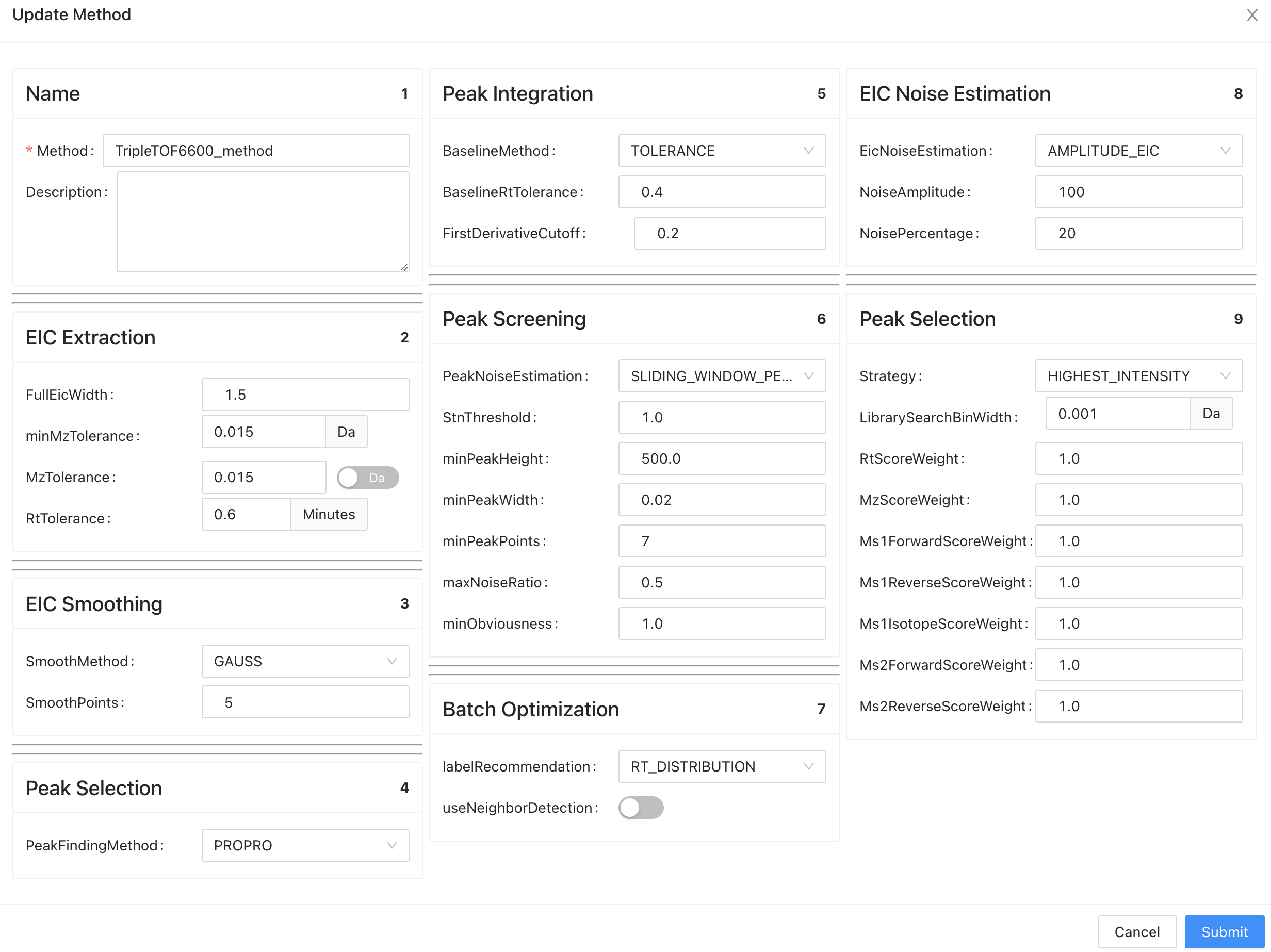
The parameters explanation is summarized below:

You can click Edit on the method page to change analysis parameters.
Suggestions:
- We recommend using different methods for data under different experimental conditions, such as different mass spectrometers.
- The optimized method should be less modified.
4.2.4 Start analysis
- Enter Project page, click
Target Analyzeat the row end.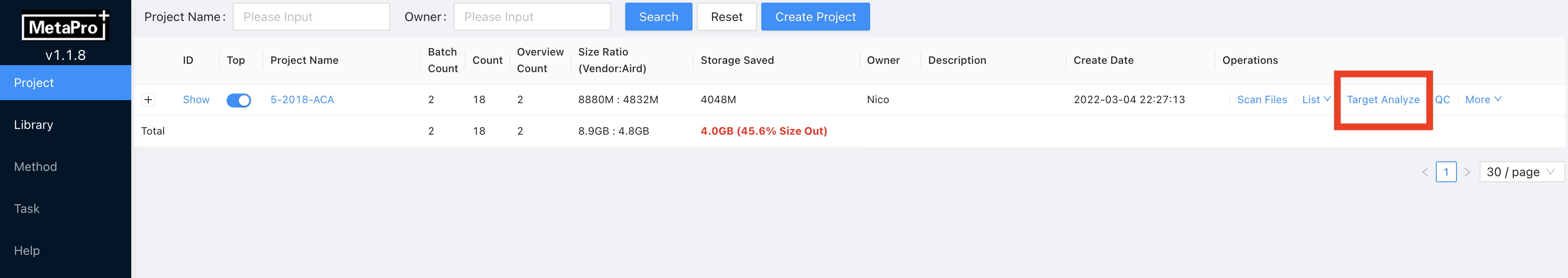
- Click
Editto change the libraries and method for batch targeted analysis.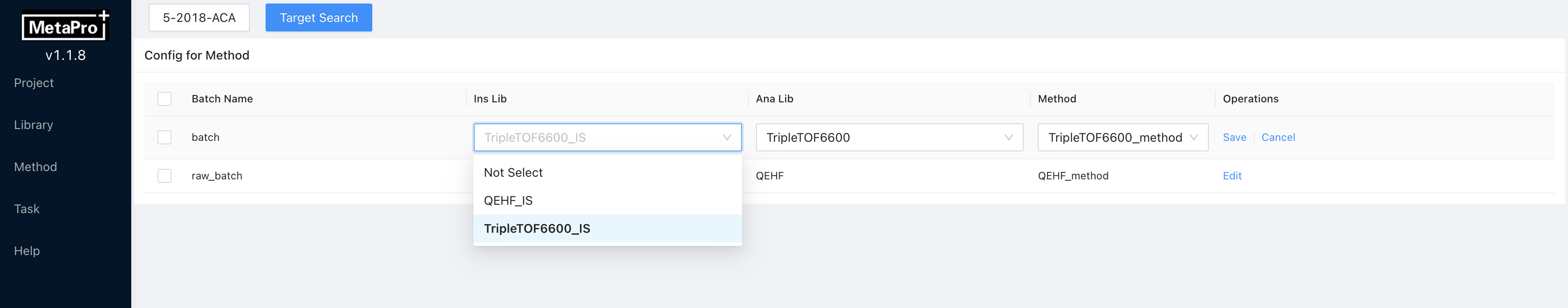 Click
Click Saveto commit changes. Multi-batch editing is also supported. You can select multiple batches by clicking on the checkboxes on the left. Then clickMore → Edit, and edit libraries and methods for multiple batches.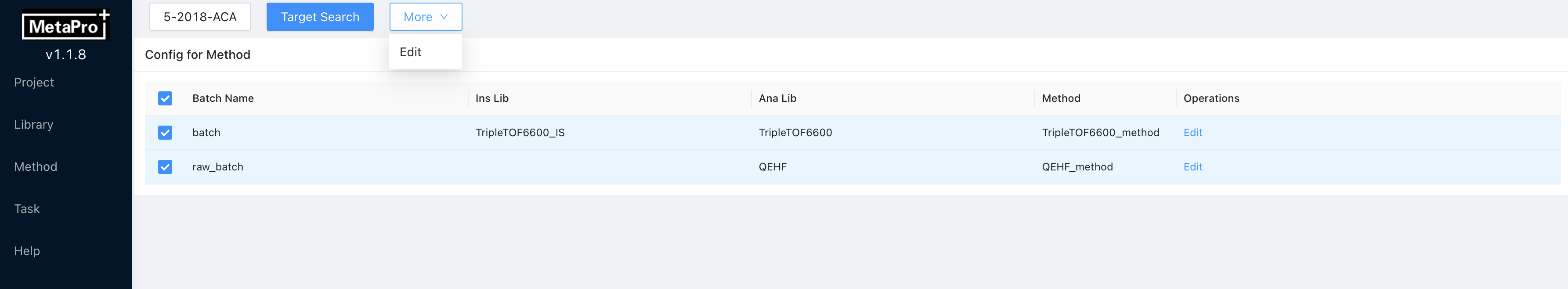
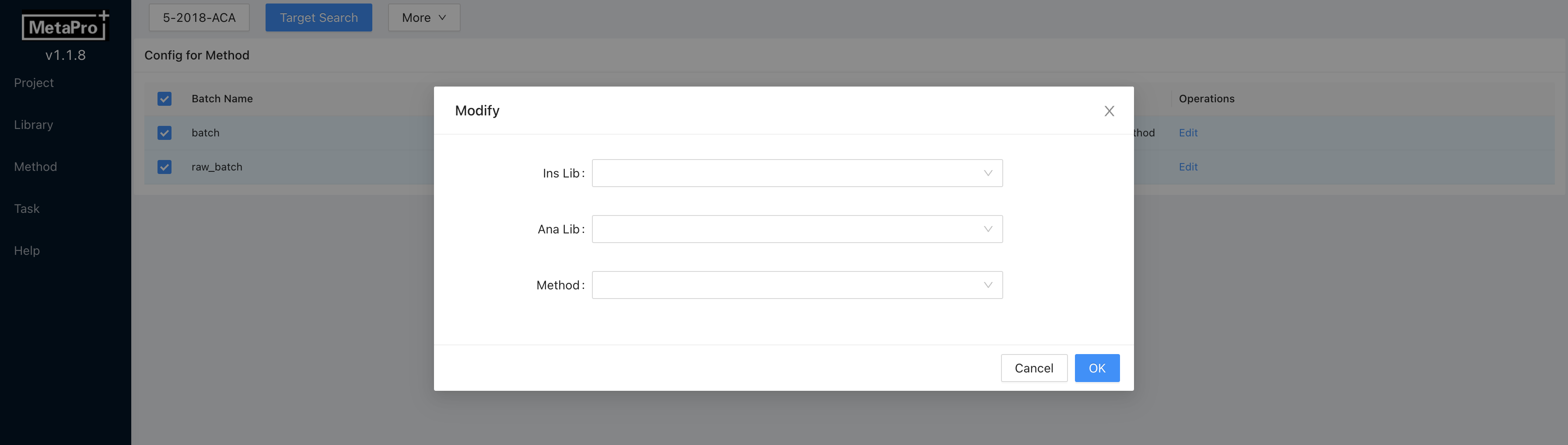
- Click
Target Searchto start a targeted analysis. You can select multiple batches and clickTarget Searchto submit multiple analysis tasks. MetaPro has a job queue and the tasks will be analyzed batch by batch. The analysis progress can be viewed at the Task page.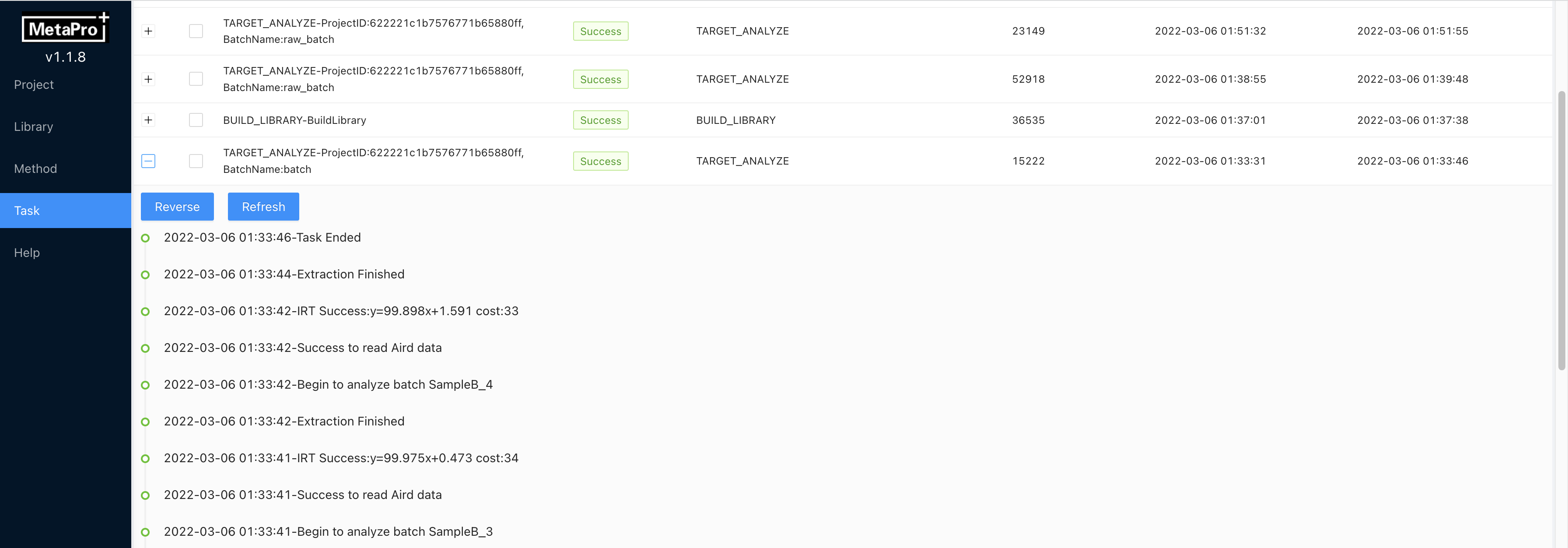 Results can be found in QC page after the analysis process is finished. During the analysis process, all MetaPro functions are not affected.
Results can be found in QC page after the analysis process is finished. During the analysis process, all MetaPro functions are not affected. - Result management.
- [View] Click
+button of project in Project page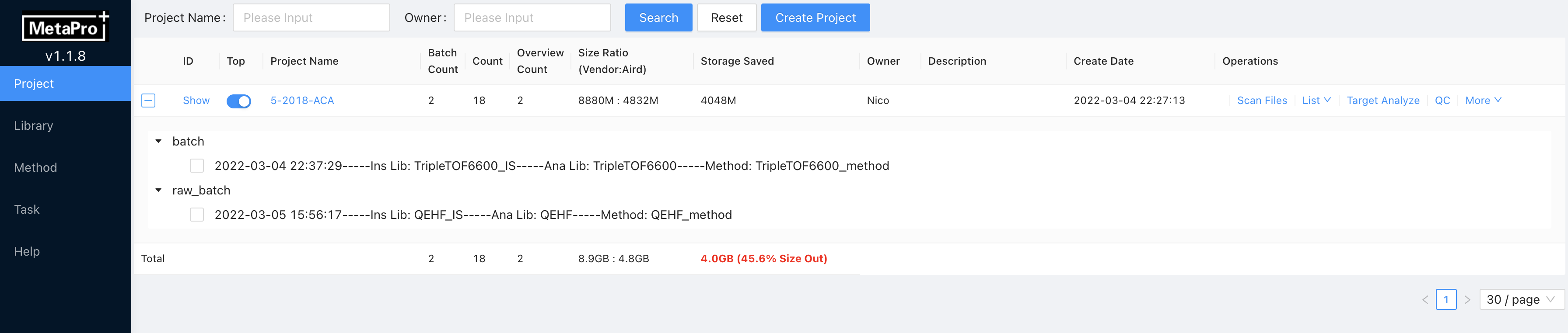
- [View & Delete] Click
List → Overview List
- [View] Click
4.3 Use batch QC inspection page for targeted analysis
4.3.1 Access the batch QC inspection page
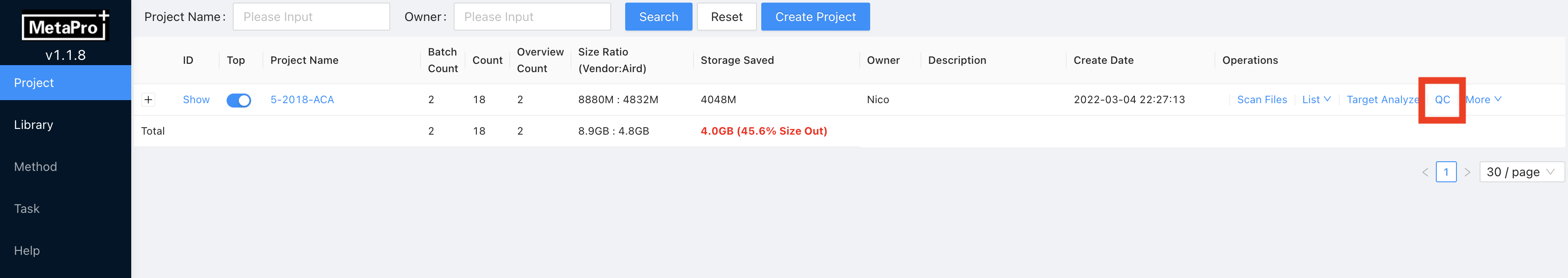
If the targeted analysis process has been finished, then you can view the quantification results on the batch inspection interface.
Enter project page and click on QC on the project that you want to view.
4.3.2 Batch Quality Control inspection interface
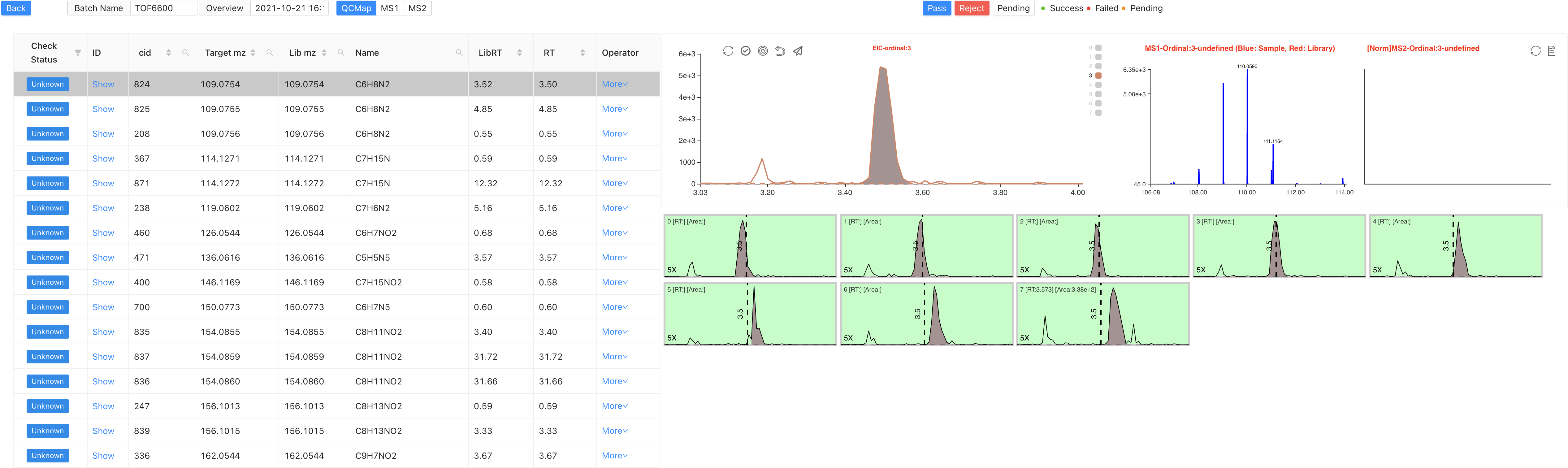
This is the main inspection page on samples in a batch. It includes different function modules as labeled.
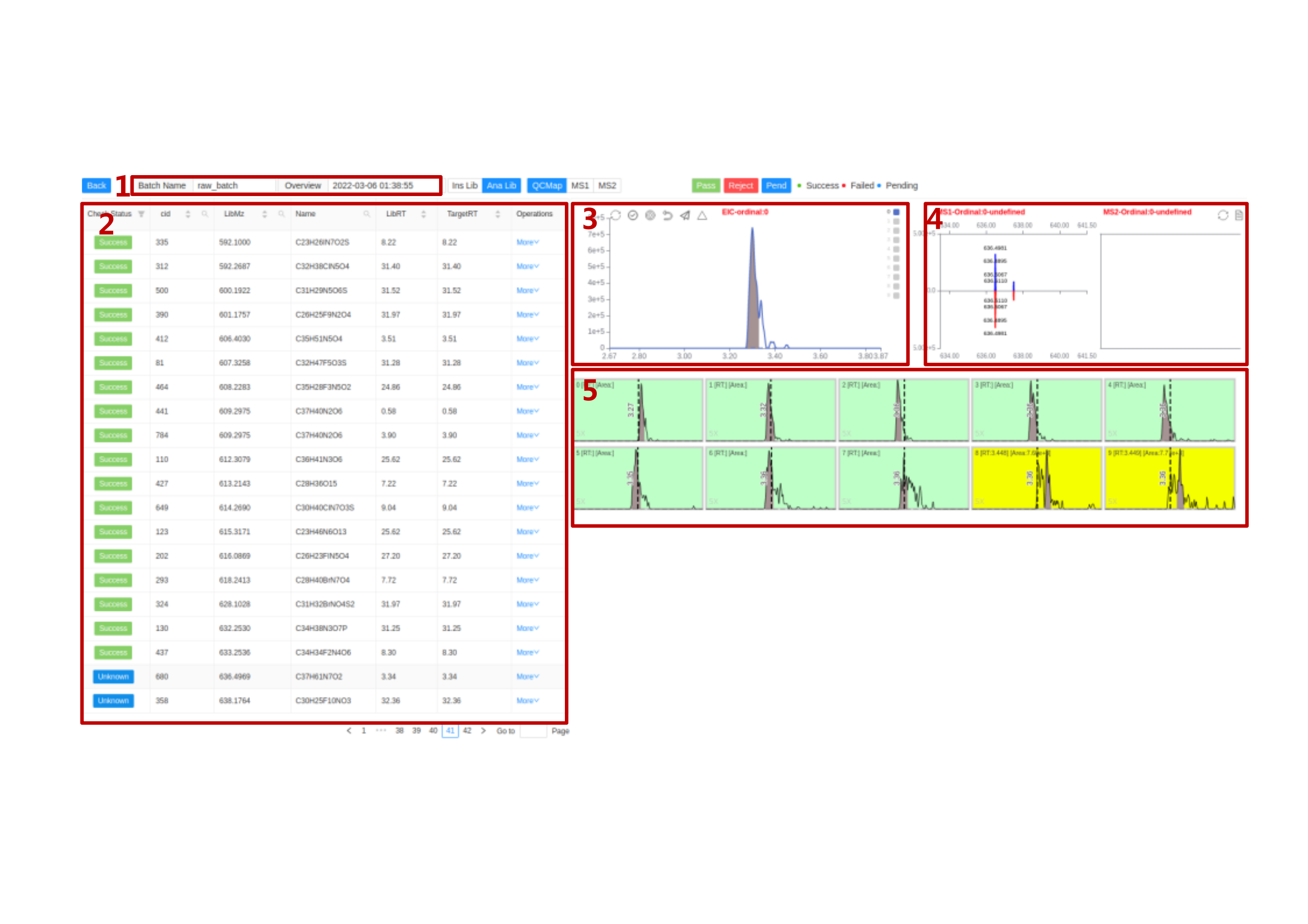
Overview switch module Click on the
Batch NameorOverviewdialog on the left top. You will see the following interface. You can change the current overview to another by clicking on a specific overview.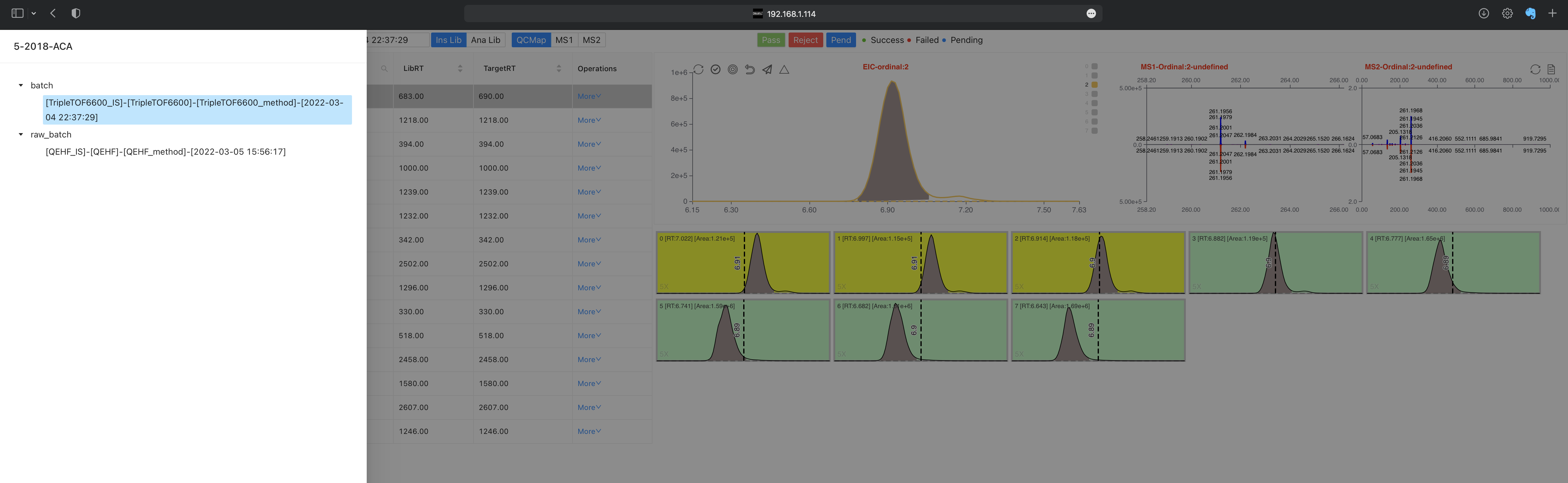
Compound switch module
Check Statusrepresents for the current check status of compounds. It has Success、Failed、Unknow status.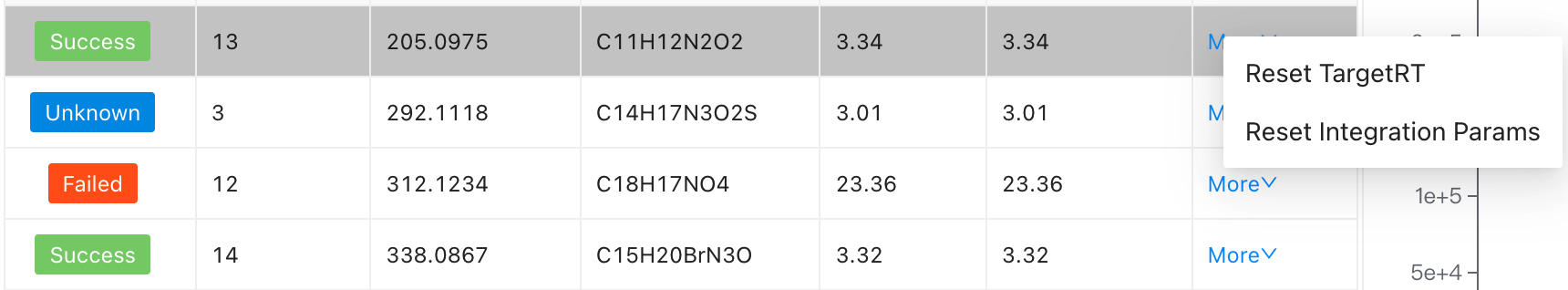 Shortcuts
Shortcuts
Arrow keys:
↑↓switch componund;→←switch pageControl the check status of the current compound,
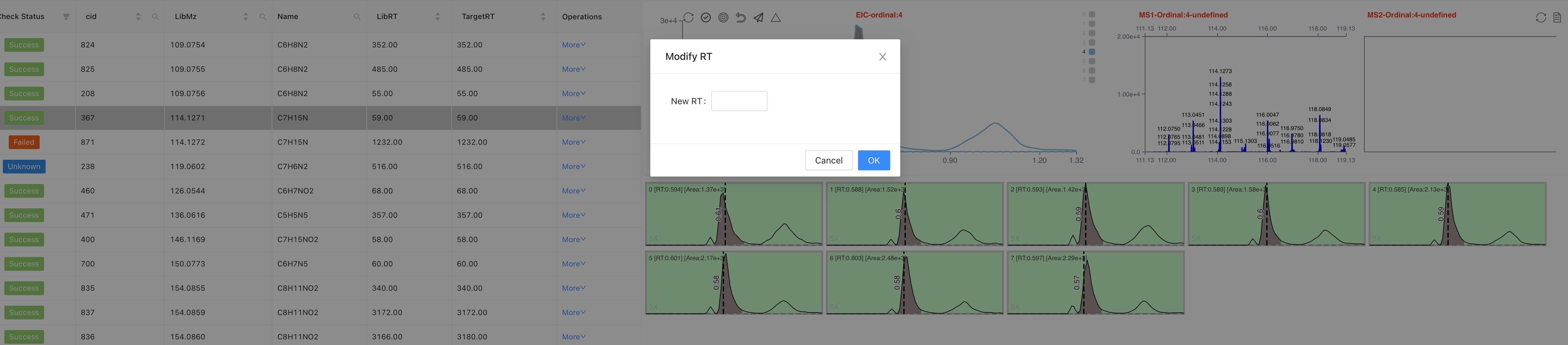
Space ␣andEnter ⏎: Success,BackSpace ⌫: FailedDouble clickon each row to change RT valueClick on the "more", users can
Reset integration paramsandReset TargetRTof a compound.Click on
Reset TargetRT
- Click on
Reset integration params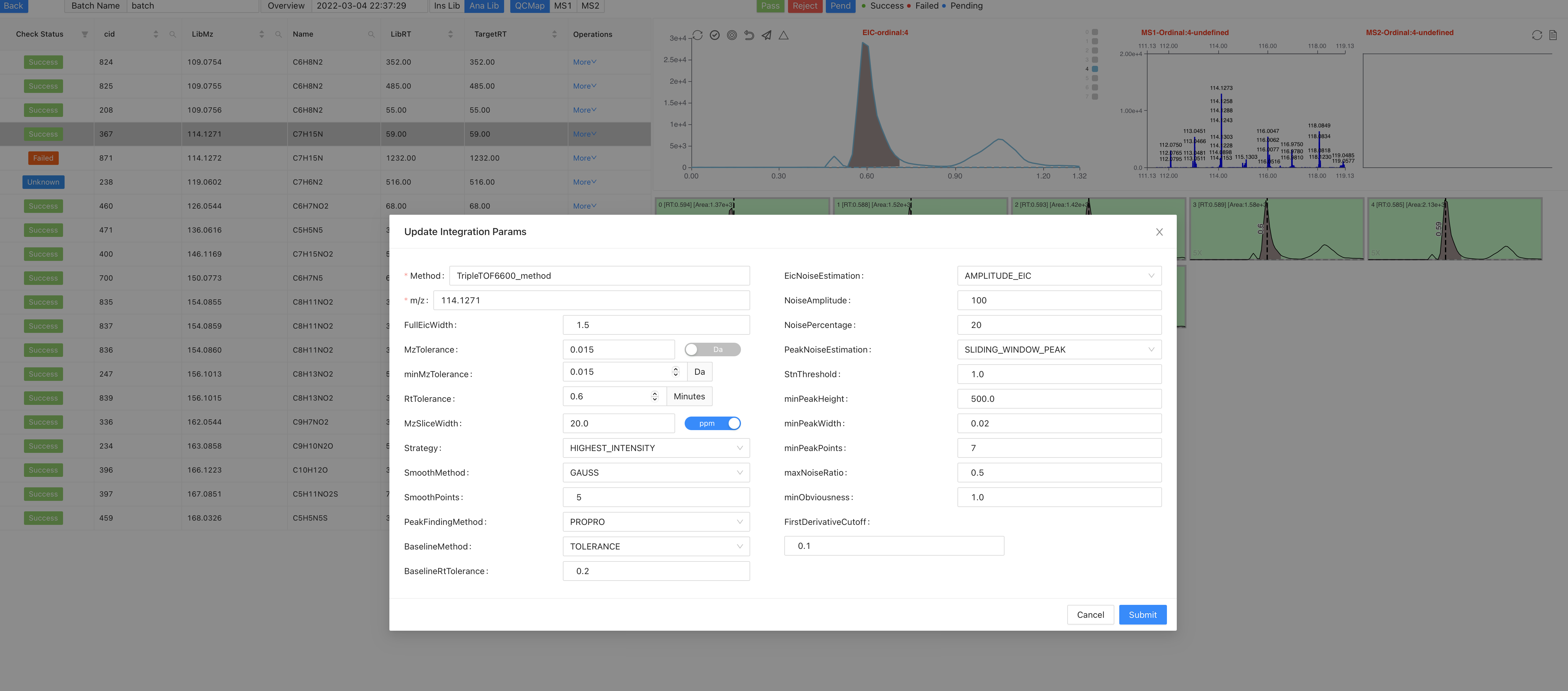
Manual inspection module The buttons on the top are:
Restore,Select All/UnSelect,Manual Integration,Reselect,Submit,Batch SubmitRestore: Restore the current module to its initial status.Select All/Unselect: View all the sample peaks in this batch/Cancel view all the peaks.Manual Integration: Enter manual integration mode on the selected peak. You can choose two data points (start, end) to modify the peak shape.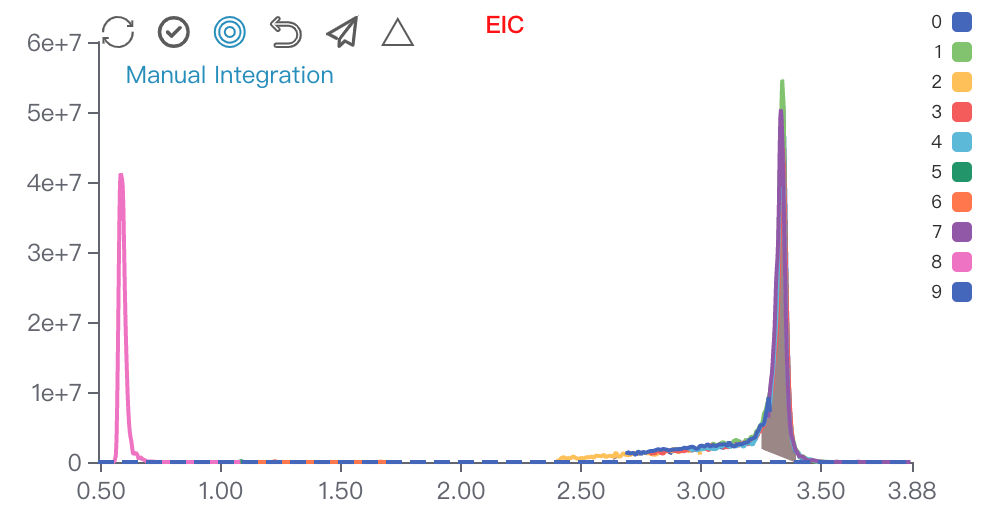
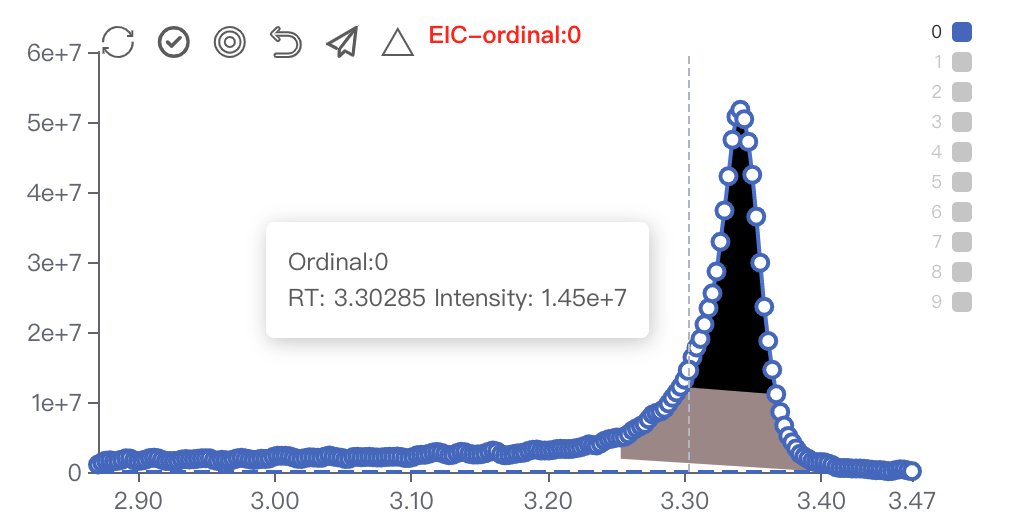
Reselect: Click this to rechoose the data points.Submit: Submit the modified peak shape to database.Batch Submit: Apply the modified peak RT range to all samples in this batch. Used for integrating multiple peaks on an EIC.Click on
Manual Integration, you can choose two data points to conduct manual integration functions. First left and then right.Spectrum viewer of
MS1andMS2This is changed according to the sample you choose on the batch inspection module.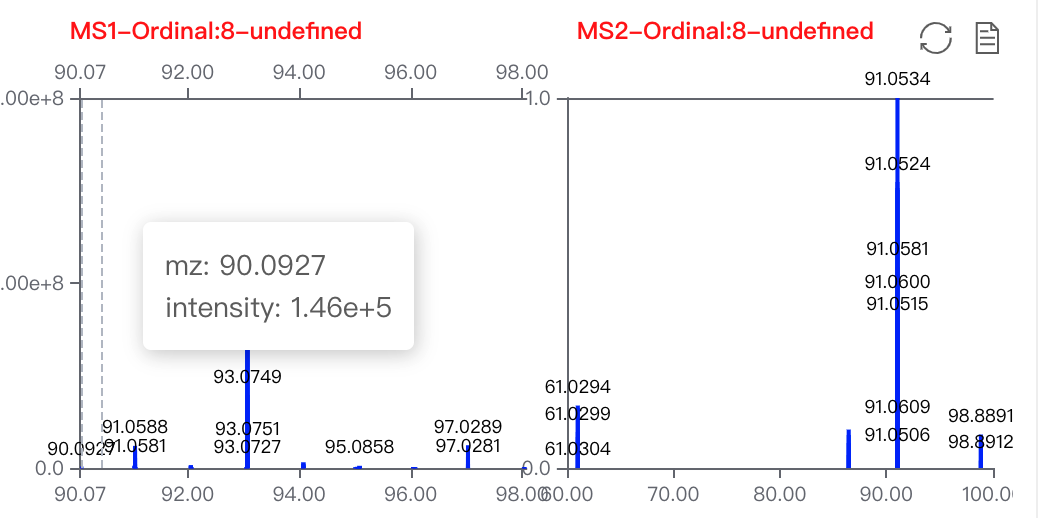 This is the Spectrum viewer towards all the
This is the Spectrum viewer towards all the MS1andMS2. Click on the pointed label to switch pages. Shortcuts
Shortcuts- keyboard
1,2,3:switch check status A:choose all the samplesESC:cancel check all
- keyboard
Peak shape batch inspection Users can choose multi-samples to change their status. Red represents for rejection. Yellow represents for unknown. Green represents for pass. The shortcuts are
1,2,3. KeyboardAis to choose all the samples. If the status of a sample is labeled in yellow or red, its quantification result area will be set to 0.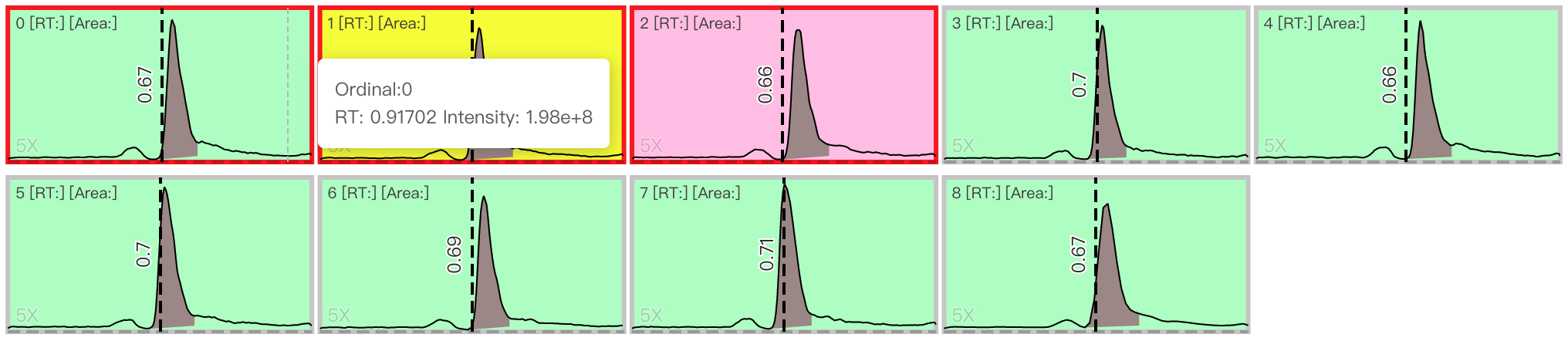
4.3.3 QC inspection interface shortcuts conclusion
Compound switch module
- Arrow keys:
↑↓switch compound;→←switch page - Control the check status of the current compound,
Space ␣andEnter ⏎: Success,BackSpace ⌫: Failed Double clickon each row to change RT value
- Arrow keys:
Peak shape batch inspection
- keyboard
1,2,3:switch check status A:choose all the samplesESC:cancel check all
- keyboard
Manual integration module
Double click:submit manual integration resultsR:refresh the current page
4.4 Spectral Library Curation
4.4.1 Build Library
Choose inspected overview to conduct library curation process on a specific project.
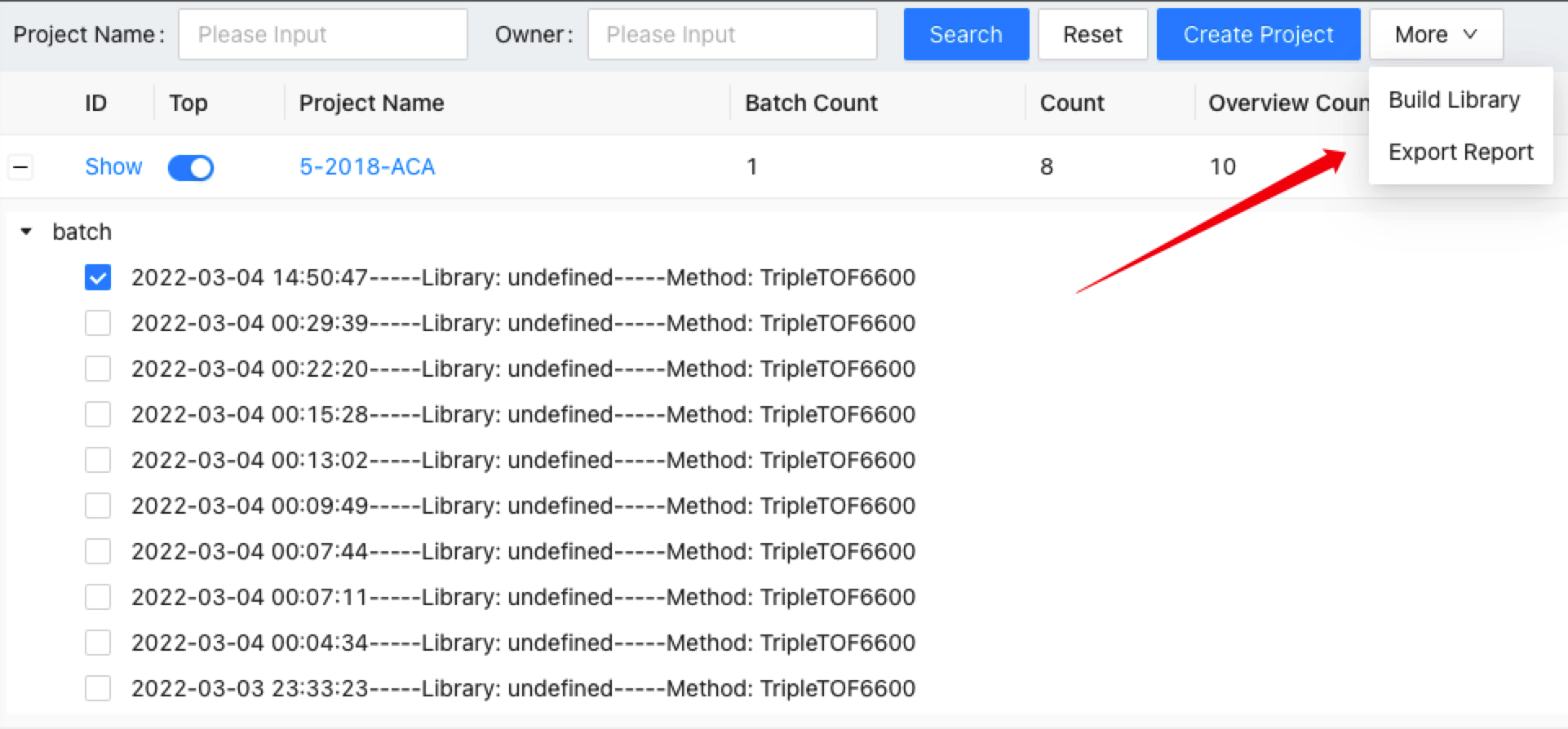
This will generate a Library Building task and you can check it on the task list. The process automatically chooses all the inspected and passed spectra to save them into the database. If you saw the task status is finished, you can conduct library QC on the curated library.
4.4.2 Library QC
After build library task has been finished on a inspected library. This library will have collected spectra waiting for inspection. Click on the QC button to view the spectra inside this library.
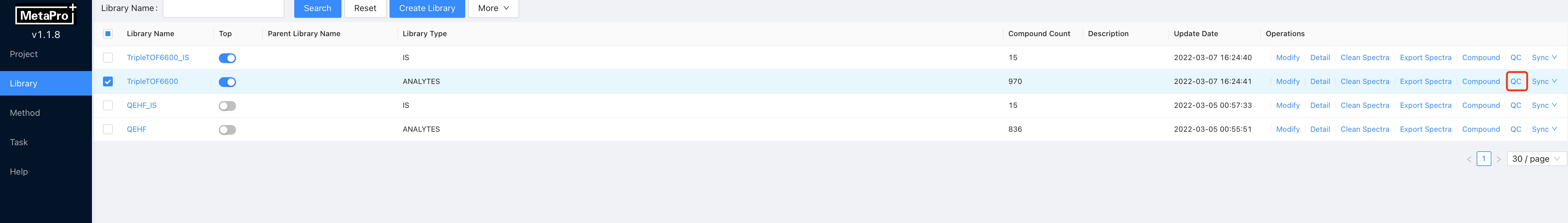
Enter the library QC interface you will see a page like that.
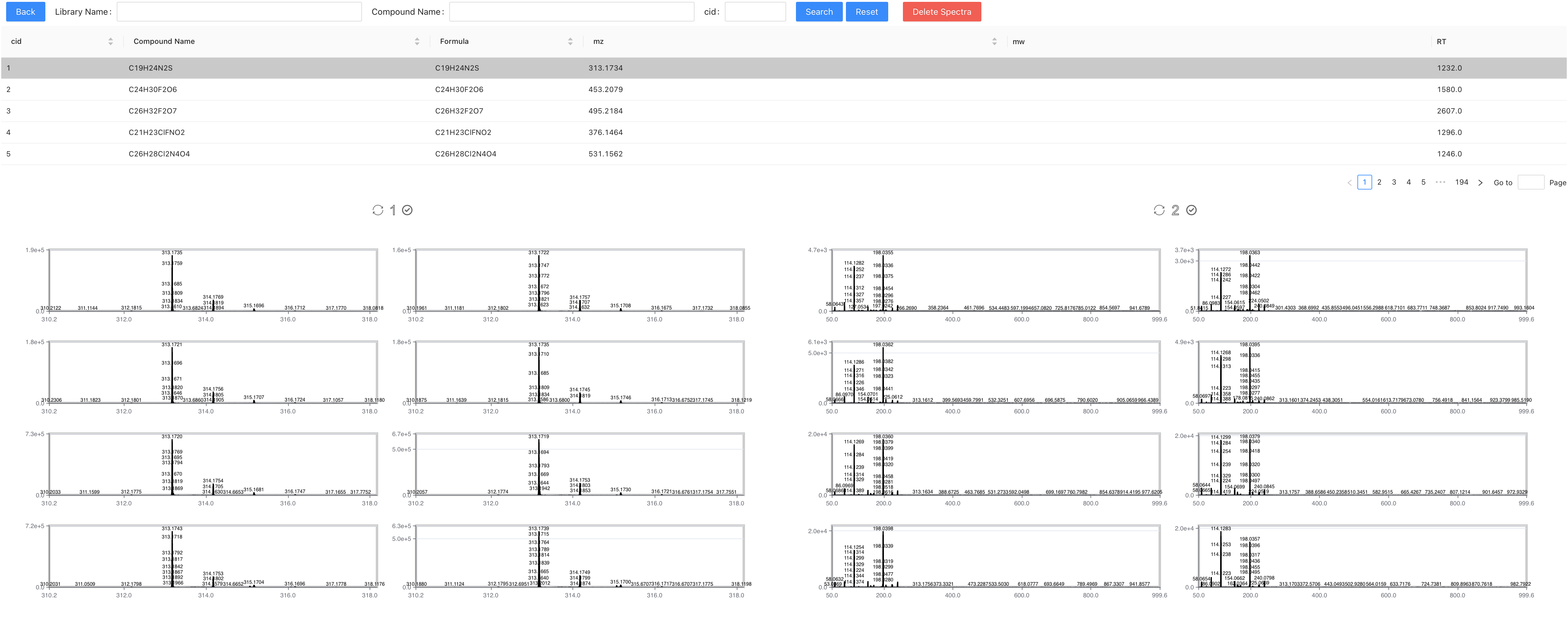
On this page you can see all the spectra collected on this compound. You can multi-choose to delete low-quality spectra.
Shortcuts:
- Arrow keys:
↑↓switch compound;→←switch page Del: Delete the selected spectra.
4.5 Export
4.5.1 Export quantification reports
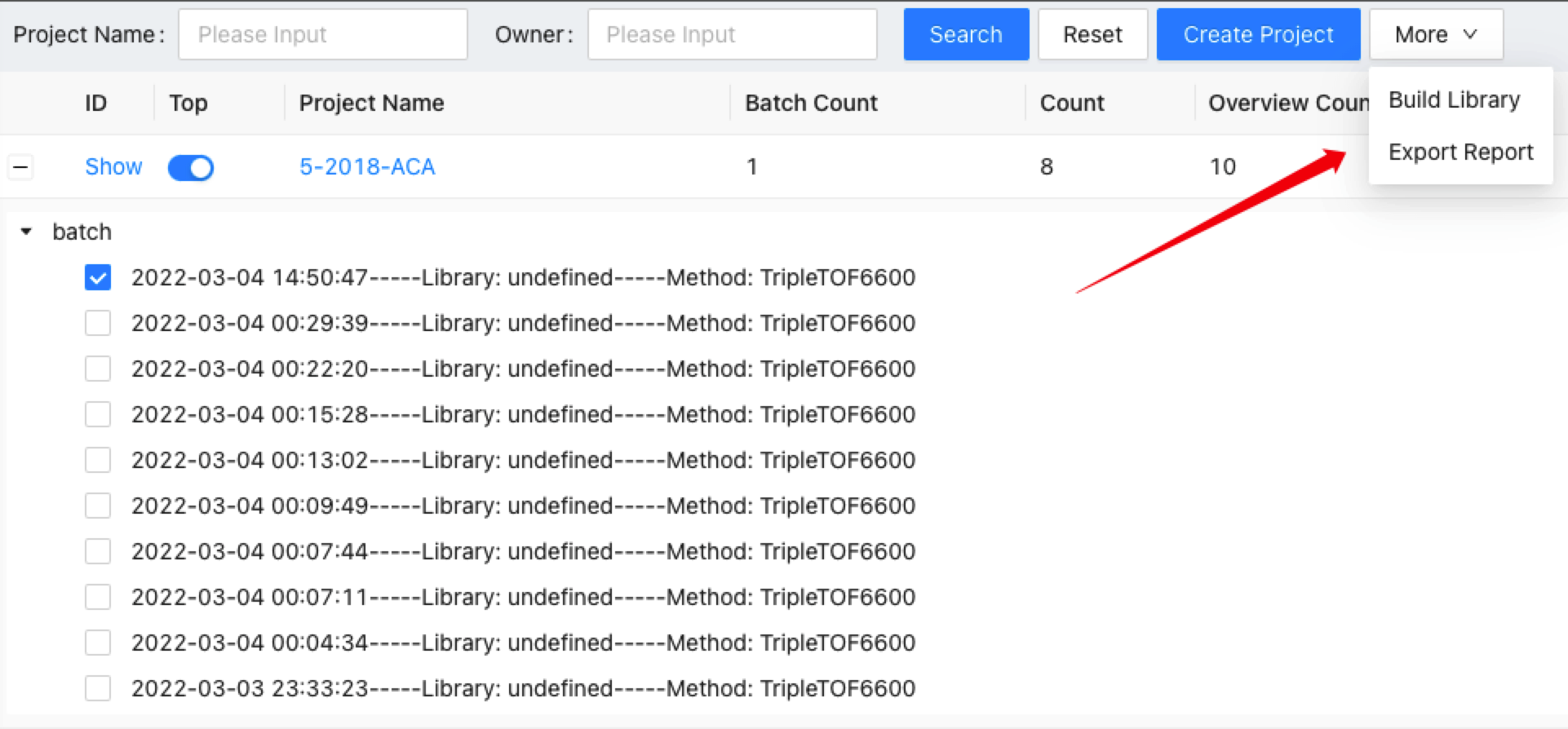
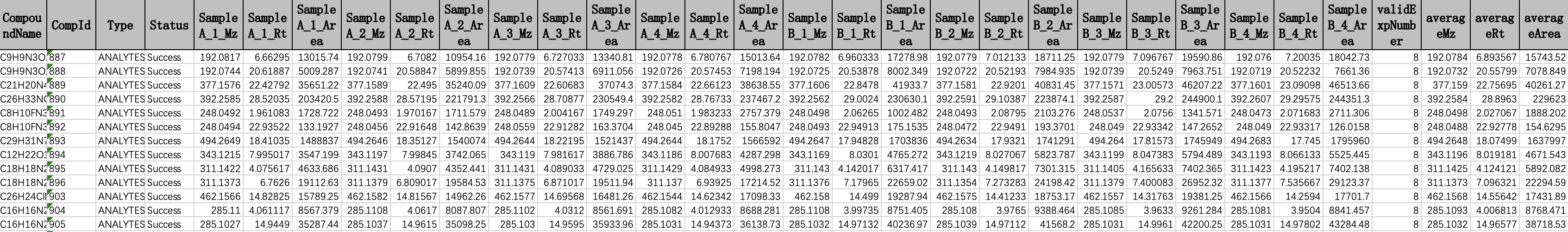
First, choose a project on the project page, click on the + to view all the overviews under this project. Select one and click the Export Report. It will let users input the output file name towards a specific overview. The quantification results of this overview will be exported into the given .xlsx file.
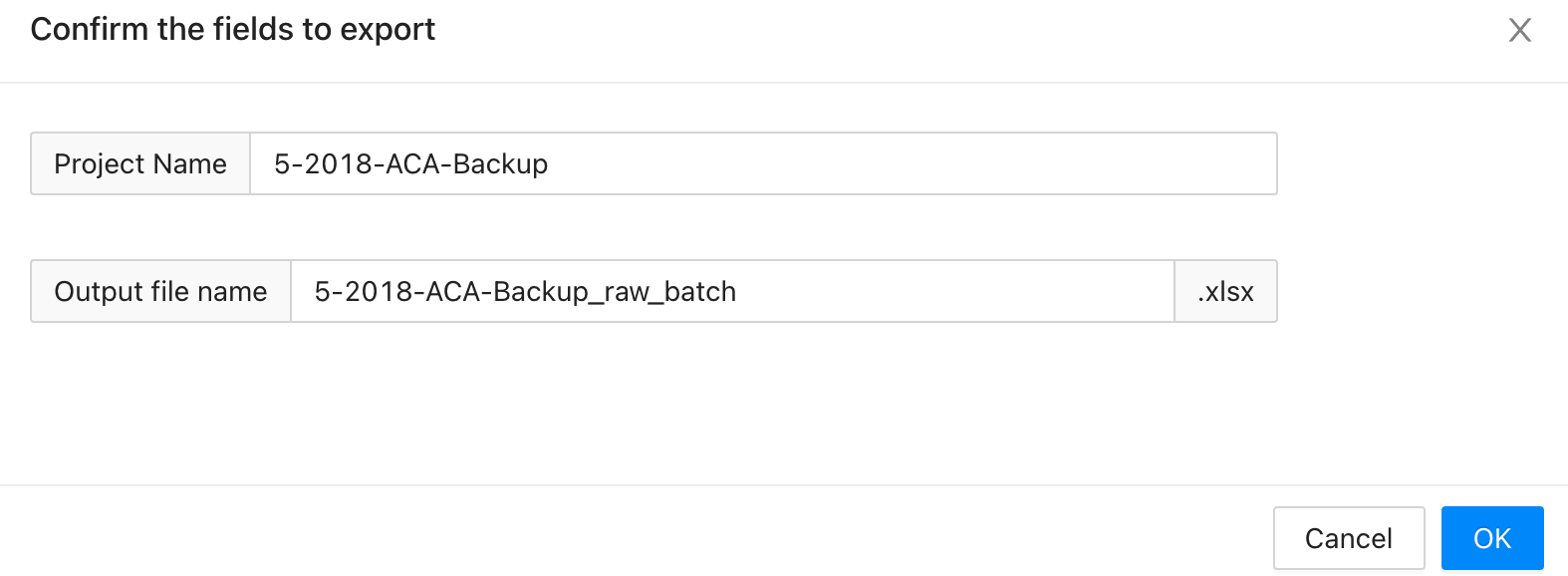
Example exported file look like
4.5.2 Export Library
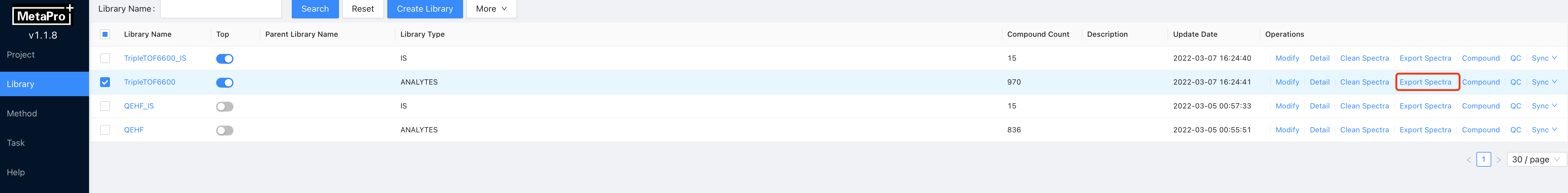
At the library page, click on Export Spectra button will let MetaPro output the curated spectra to the given export file path according to the compounds. The spectra are stored in txt format with the first column representing for m/z and the second column representing for intensity.
Supplementary
Performance Optimization
- NOTE: This step has relatively complicated operations. Users who need to perform high-throughput inspection analysis on the server can perform performance optimization according to the following description.
MetaPro uses in-memory caching technology to speed up the analysis of MS data.
In the manual inspection process, each time the user changes the integration parameters, MetaPro needs to re-analyze all files in the current batch. Most of the analysis time is spent reading the files, about 1 secondper file on the tutorial dataset. If a user is inspecting a batch with 10 data filesand wants to change the integration parameter of a compound, without memory caching, it will take 10 seconds to load files and re-analysis the compound within 1 second. MetaPro accelerates repeated reading of files through caching technology. In the case of sufficient memory, MetaPro can load the file from disk only once when recalculating the first compound, save the file reading time for the inspection of other compounds, and achieve efficient batch manual quality control.
Considering the performance of most users' computers, we configure a small amount of cache in the default installation. Users can change the cache configuration themselves to achieve faster response and higher inspection efficiency.
The configuration file is located at INSTALLATION_PATH/libs/config/ehcache.xml
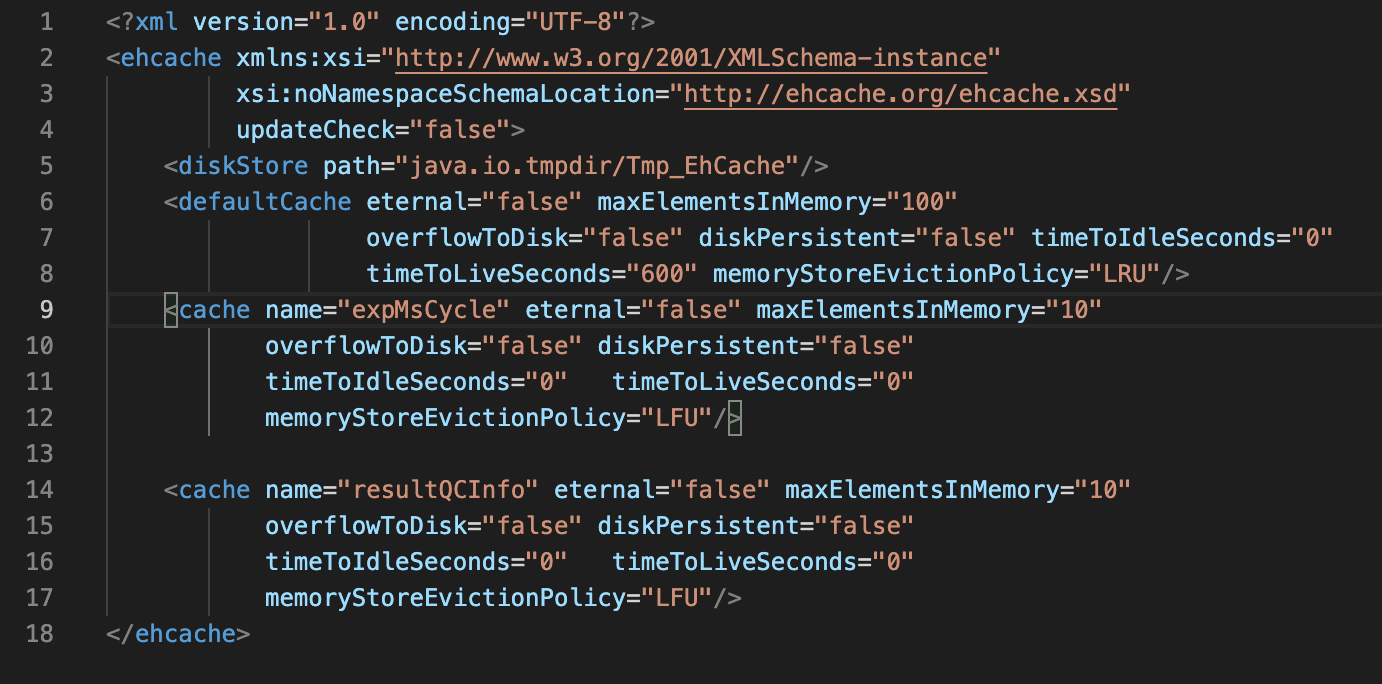
Users can moderately increase the value of maxElementsInMemory at line 9, according to the computer memory size. This value means the max number of cached files. If the memory size is big enough, maxElementsInMemory should be larger than the number of files in each batch. If not, try separating data into multiple batches to avoid disk reading.
Save the config file, restart MetaPro and enjoy.
